Biopython version 1.86
© 2025. All rights reserved.
© 2025. All rights reserved.
This page describes the Contig class used in Bio.Sequencing.Ace to hold all of the information about a single contig record in an Ace file.
A contig is a set of overlapping sequences used to generate a consensus sequence for a given region of a genome. Ace files are usually used to store contigs (that is a consensus sequence and the DNA sequences that are used to generate it) created with phrap or as part of 454 sequencing projects. Biopython’s ace contig class follows the the structure ace file format closely so if you are not familiar with these files (a description of the format can be found under the heading “ACE FILE FORMAT” in the consed readme) it might represent a steep learning curve - the diagram below gives an overview of how some of the most important information is stored.
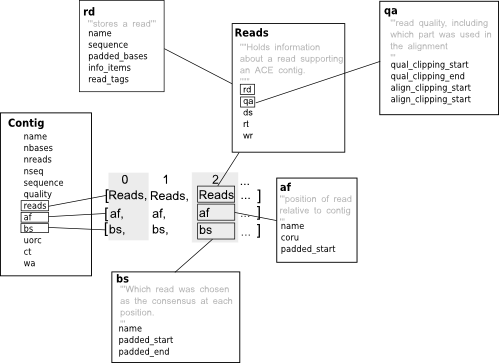
As we’ve said contigs can be used to represent a sequence corresponding to some part of a genome (or transcripome or whatever -ome it is that you are studying). Treating contigs as sequences is pretty straight forward, using the contig class you can do it this way:
#Start by parsing a file to get some contigs
>>>from Bio.Sequencing import Ace
>>>ace_gen = Ace.parse(open("contig1.ace", 'r')) # from Tests/Ace/contig1.ace
>>>contig = ace_gen.next()
# Now do some stuff with them
# what's our contig called
>>> contig.name
'Contig1'
# just the consensus sequence
>>>contig.sequence[:78]
'aatacgGGATTGCCCTAGTAACGGCGAGTGAAGCGGCAACAGCTCAAATTTGAAATCTGGCCCCCCGGCCCGAGTTGT'
# assembler designated quality for each base in the consensus
>>> contig.quality[:20]
[0, 0, 0, 0, 0, 0, 22, 23, 25, 28, 34, 47, 61, 46, 39, 34, 34, 30, 30, 31]
Ace files are also supported by the SeqIO module
>>>from Bio import SeqIO
>>>ace_gen2 = SeqIO.parse(open("contig1.ace", 'r'), 'ace')
>>>new_contig = ace_gen2.next()
>>>print(new_contig)
ID: Contig1
Name: Contig1
Description: <unknown description>
Number of features: 0
Per letter annotation for: phred_quality
Seq('aatacgGGATTGCCCTAGTAACGGCGAGTGAAGCGGCAACAGCTCAAATTTGAA...tac', Gapped(DNAAlphabet(), '-'))
# You can do all the normal SeqIO tricks like file conversion
>>>sequences = SeqIO.parse(open("contig1.ace", 'r'), 'ace')
>>>SeqIO.write(sequences, open("ace1_consesus.fasta", "w"), "fasta")
Ace files also contain information on the reads that support the consensus sequence. Its possible to treat each contig as a generic alignment using this cookbook entry (there has been a discussion on bringing a function like this into AlignIO here, if you have thoughts with how best to do this then please share them on the list). It’s also possible to get a bunch of properties for each read from two lists in the contig class, ‘reads’ and ‘af’
# using the contig we read into memory above
# get the name and the sequence of each read in the contig
for i in range(len(contig.reads)):
print("read %s: %s" % (contig.reads[i].rd.name, contig.reads[i].rd.sequence[:50]))
# ...
# read BL060c3-LR5.g.ab1: tagcgaggaaagaacccaacaGgaTTGCCCTAGTAACGGCGAGTGAAGCG
# read BL060c3-LR0R.b.ab1: aatacgGGATTGCCCTagtaacGGCGAGTGAAGCGGCAACAGCTCAAATT
# the 'qa' property of each read contains the reads quality clipping
for i in range(len(contig.reads)):
print(
"for read %s newbler decided bases %i:%i where good enough to "
"include in the consensus"
% (
contig.reads[i].rd.name,
contig.reads[i].qa.qual_clipping_start,
contig.reads[i].qa.qual_clipping_end,
)
)
# ...
# for read BL060c3-LR5.g.ab1 newbler decided bases 80:853 where good enough to include in the consensus
# for read BL060c3-LR0R.b.ab1 newbler decided bases 7:778 where good enough to include in the consensus
# the 'af' list has information on the read relative to the consensus it supports
for i in range(len(contig.reads)):
print(
"the first base in %s aligns with base %i in the consensus"
% (contig.reads[i].rd.name, contig.af[i].padded_start)
)
# ...
# the first base in BL060c3-LR5.g.ab1 aligns with base -14 in the consensus
# the first base in BL060c3-LR0R.b.ab1 aligns with base 1 in the consensus